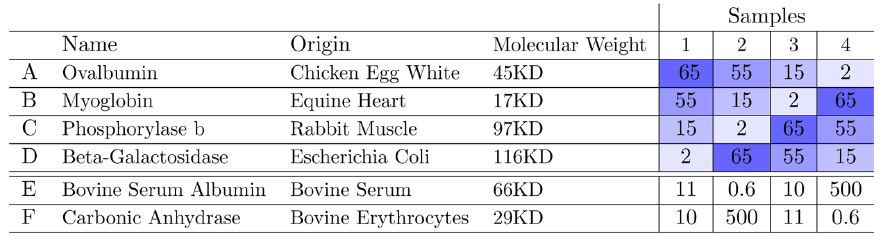
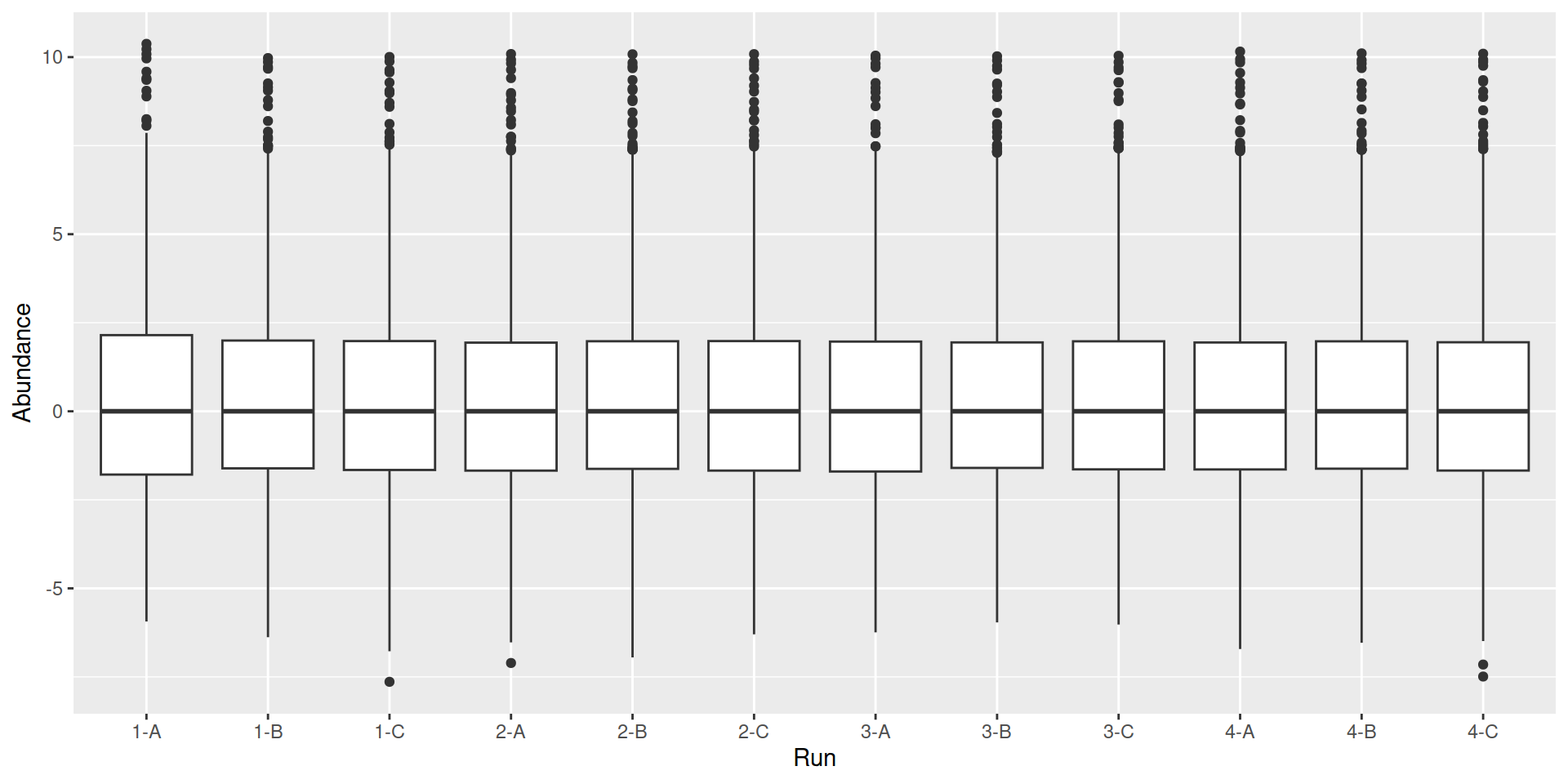
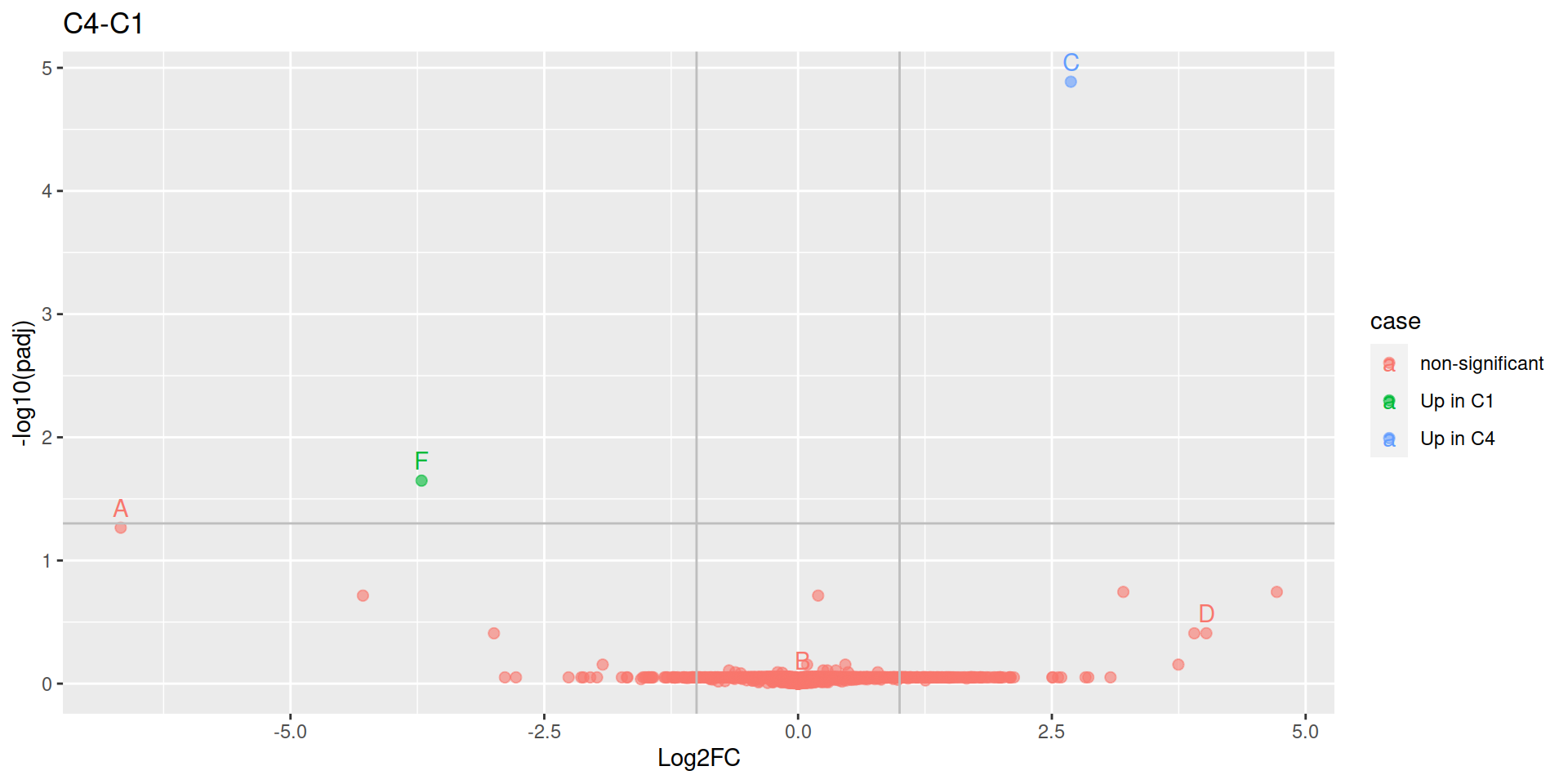
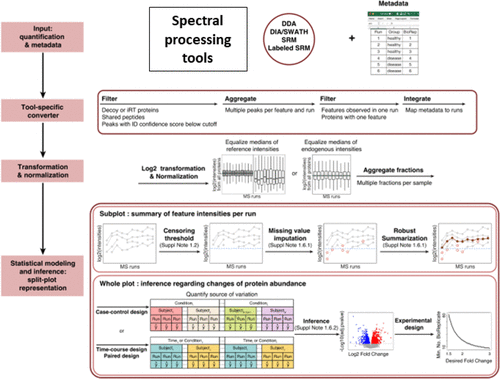
Choi, Meena, Zeynep F. Eren-Dogu, Christopher Colangelo, John Cottrell, Michael R. Hoopmann, Eugene A. Kapp, Sangtae Kim, et al. 2017.
“ABRF Proteome Informatics Research Group (iPRG) 2015 Study: Detection of Differentially Abundant Proteins in Label-Free Quantitative LC–MS/MS Experiments.” Journal of Proteome Research 16 (2): 945–57.
https://doi.org/10.1021/acs.jproteome.6b00881.
Jones, Jeff, Elliot J. MacKrell, Ting-Yu Wang, Brett Lomenick, Michael L. Roukes, and Tsui-Fen Chou. 2023.
“Tidyproteomics: An Open-Source r Package and Data Object for Quantitative Proteomics Post Analysis and Visualization.” BMC Bioinformatics 24 (1): 239.
https://doi.org/10.1186/s12859-023-05360-7.
Kohler, Devon, Mateusz Staniak, Tsung-Heng Tsai, Ting Huang, Nicholas Shulman, Oliver M. Bernhardt, Brendan X. MacLean, et al. 2023.
“MSstats Version 4.0: Statistical Analyses of Quantitative Mass Spectrometry-Based Proteomic Experiments with Chromatography-Based Quantification at Scale.” Journal of Proteome Research 22 (5): 1466–82.
https://doi.org/10.1021/acs.jproteome.2c00834.
Lazar, Cosmin, Laurent Gatto, Myriam Ferro, Christophe Bruley, and Thomas Burger. 2016.
“Accounting for the Multiple Natures of Missing Values in Label-Free Quantitative Proteomics Data Sets to Compare Imputation Strategies.” Journal of Proteome Research 15 (4): 1116–25.
https://doi.org/10.1021/acs.jproteome.5b00981.